Ontology Synthesis from Book Indices
keywords: AI, Reasoning, Ontologies
Introduction
A few years back I needed an ontology for biological processes. I could not find anything that fit my particular needs or that even worked as a good starting place. So, I picked up a couple of books that seemed to be relevant. As usual, I scanned the table of contents and flipped to the index. There the beginnings of a solution stared back at me.
I already knew that an extremely valuable kind of resource for the synthesis of ontological structure are journal or book indices. Clearly, the entries in an index are indicative of those concepts that were deemed important by the index creator. However, the structure of indices and inconsistencies make the extraction of concepts and relationships very difficult. The goal of this note is to describe some of the most salient puzzles that must be solved to enable reasonably accurate extraction.
Indices are one of three kinds of sources for automated ontology creation, the other two being source ontologies which may be modified and merged using SETL techniques and information extraction from text using NLP techniques. The advantage of using indices is that they will tend to reflect the application domain"s terminology at a reasonable level of abstraction and granularity and that extraction from indices is expected to have a lower error rate than extraction from free text. Of course, indices do not contain many of the kinds of relationships that we would expect to extract from free text.
This note addresses the two principle processes that must be used to for ontology synthesis from indices. The first is the conversion of source documents to a form that is appropriately structured for extraction. This process converts PDF to text, and then parses the text to create tree-like data structures that capture the semantics that is expressed through indentation and other elemental clues for index entries.
The second process is the extraction of concepts (in general common names and named entities) and relationships from the tree-like data structures.
I am unaware of any directly similar or analogous prior art for ontology synthesis from indices other than some extraction processes that Arity and Headspace Sprockets have created for other kinds of complex textual documents.
Process 1: Document Parsing
A typical PDF index (this one from MCB in 1995) looks like the following (cutting the top and the bottom of a typical page):

We use the state of the art tool pdftotext (http://www.foolabs.com/xpdf/home.html).
Characters are converted to their nearest equivalents and indentation is approximated by series of spaces. A number of problems arise:
- Many fonts are subject to conversion errors, including but not limited to italicized fonts and stylized fonts such as often used for headers. For example, "in vitro" sometimes converts into "m vitro". There does not seem to be any solution to this problem.
- Greek letters (e.g. $\beta$) show up as other Latin1 chars.
- Some words get inappropriately spaced. For example, the word "See" may sometimes be converted to "S e e".
- Some words that have doubled consonants do not convert correctly. For example, "embedding" may be converted to "embeYing".
- Spacing of columns often is inconsistent apparently because of errors in calculating the number of padding spaces to be added. Some of these errors seem to be greater than would be expected by merely going from variable to fixed pitch fonts.
There is no known solution to these problems based on settable parameters or the use of other programs. The document parser includes some strategies for error repair and for robust processing of indentation.
Document parsing must strip headers and footers. This requires an analysis of the document to determine how to recognize which lines are indeed able to be discarded from further processing for each page.
Then, for each page, each line must be processed column-wise. This process requires an analysis pass which will determine the number of columns (typically but not necessarily two) and the amount of indentation that is used to demark columns and to demark continuations and sub-entries. Usually continuation lines are indented further than sub-items.
The analysis and interpretation of indentation is very complex and must account for continuations (entries longer than a single line), for parenthetical comments such as "see also -", for page information (which may be omitted, contain single pages, page ranges, or comma separated combinations), and for sub-items. Rather pathologically, I have found that each page may have different indentation characteristics introduced by the PDF to text conversion process. Each page is analyzed separately. This introduces some complexity when entries span more than one page.
To illustrate, we may have the following lines (taken from the PDF example above and exactly as produced by pdftotext).
Cosmids labeling, 23, 25
ACeDB, 595,596 DEB, see Diepoxybutane
cloning, 470-471, 552 deb-I gene, 38, 85, 90
contigs, 536, 539 Deficiencies, genetic balancers, 158
DNA, 347, 349, 455 Degenerate oligonucleotide primed PCR, 352-353
miniprep, 543-544 Degenerate primers, 353
preparation, 552-553 Degenerins, 248
fingerprinting, 554 deg-1 gene, 248
finishing, 561-568 DEO, see Diepoxyoctane
growth and handling, 541 Detectional sequencing, 578-58 1
sonication, 555 Developmental pathwaysIf we focus on processing the items in the first column we can see two levels of indentation. (The second column has examples of "see-" but neither has continuation lines). An intermediate representation of the text is the following:
l(0,"Cosmids",upper(67),lower(115))
l(3,"ACeDB, 595,596",upper(65),digit(54))
l(3,"cloning, 470-471, 552",lower(99),digit(50))
l(3,"contigs, 536, 539",lower(99),digit(57))
l(3,"DNA, 347, 349, 455",upper(68),digit(53))
l(6,"miniprep, 543-544",lower(109),digit(52))
l(6,"preparation, 552-553",lower(112),digit(51))
l(3,"fingerprinting, 554",lower(102),digit(52))
l(3,"finishing, 561-568",lower(102),digit(56))
l(3,"growth and handling, 541",lower(103),digit(49))
l(3,"sonication, 555",lower(115),digit(53))The first number (0, 3, or 6) represents the degree of indentation). The interpretation of this sequence is a set of Prolog structures of the following form:
i([
"Cosmids"
],
"",
"").
i([
"Cosmids",
"ACeDB"
],
"595,596",
"").
i([
"Cosmids",
cloning
],
"470-471, 552",
"").
i([
"Cosmids",
contigs
],
"536, 539",
"").
i([
"Cosmids",
"DNA"
],
"347, 349, 455",
"").
i([
"Cosmids",
"DNA",
miniprep
],
"543-544",
"").
i([
"Cosmids",
"DNA",
preparation
],
"552-553",
"").
i([
"Cosmids",
fingerprinting
],
"554",
"").(And many others that I have not reproduced here).
From these structures things are already looking up. We have one or more strings, suitably separated that reflect the semantics implied by the indentation, where the entire "path" is provided for each item, e.g., ["Cosmids", "DNA", preparation]. This is a compound construction of length three.
These structures are the primary output of the first process, the parsing of the document structure for indices.
Process 2: Semantic Interpretation
Using the structures emitted from the document parsing process, we are in a position to identify concepts and relationships. This is again a very complex process involving the semantics on nominal compounding somewhat modified for the vagaries of indices. Structures also may be viewed as instances of bare argument ellipsis.
To illustrate with an example, we can look at another set of entries in the same PDF:
Antibodies
monoclonal antibodies, 377-379, 382-383
to nematode antigens, 377-383
polyclonal antibodies, 377, 379-380
staining, 316-317, 365-366, 371-372, 374-376An analysis should include essentially the following:
- Antibodies - a concept
- monoclonal antibodies
- a concept
- subsumed by "antibodies" (the term "monoclonal antibodies" completely contains its parent term, a very strong indicator)
- "to nematode antigens" is a PP which implies
- "nematode antigens" is a concept
- "antibodies to nematode antigens" is a concept which is subsumed by "antibodies"
- a relationship exists between "antibodies to nematode antigens" and "nematode antigens" where the relationship type is unspecified.
- polyclonal antibodies
- a concept
- subsumed by "antibodies"
- staining - where "Antibodies staining" is a concept and using a bootstrap ontology may that contains "staining" (presumably a method) implies an assertion that "antibodies staining" is subsumed by "staining"
- some relationship exists between "antibodies" and "antibodies staining" where we can by using a bootstrap ontology that contains "staining" and "antibodies" infer a possible type.
The most important points of this example are:
- Having good quality bootstrap ontologies (those which contain some conceptual structure used as a priori knowledge) can enable many more inferences than can be made without such ontologies
- The syntactic form of compound constructions is a key indicator of the semantics of the compound. A bootstrap ontology enables the domain of concepts to be determined (often). Together, relationships may be inferred and often typed.
- In principle, the semantic interpretation process can be used to extend a bootstrap ontology in such a way that the extended bootstrap ontology may be used to for an additional pass on the input: bootstrapping.
The syntax and semantics of compounds is interesting and bears some additional discussion. The following has been adapted from Ray Jackendoff"s "Compounding in the Parallel Architecture and Conceptual Semantics". These schematic forms are for binary compounds. We commonly encounter deeper compounds in indices. The top-down nature of these compounds allows us to reduce all processing to sequences of binary compound constructions.
The following defines the formal N-N compound schemata (or constructions) that we encounter:
Argument schema: [$N_1$ $N_2$] = [$Y_2 (\dots, X_1, \dots)$] $\qquad$ ‘a $N_2$ by/of/... $N_1$’
Modifier schema: [$N_1$ $N_2$] = [${Y_2}^ \forall$; [F($\dots, X_1, \dots, \forall, \dots$)]] $\qquad$ ‘an $N_2$ such that F is true of $N_1$ and $N_2$’
Below is a list of the (most prominent) basic functions for English compounds, with examples. (This list is Jackendoff"s but is similar to those that have been produced by others). With one exception, these seem rather plausible as functions that are readily available pragmatically. (Reminder: X is the meaning of $N_1$, Y is the meaning of $N_2$, except in the last two cases.)
• CLASSIFY ($X_1$, $Y_2$), ‘$N_1$ classifies $N_2$’: beta cell, X-ray. This is the loosest possible relation, in which the meaning of $N_1$ plays only a classificatory role.
• $Y_2$($X_1$), ‘(a/the) $N_2$ of/by $N_1$’: sea level, union member, wavelength, hairstyle, helicopter attack, tooth decay. This is the argument schema. It is sometimes reversible, with the extra coercion shown in (21b): $X_1$($Y_2$), ‘an $N_2$ that $N_1$’s things’: attack helicopter, curling iron, guard dog; also ‘an $N_2$ that people $N_1$’: chewing gum, drinking water.
• BOTH ($X_1$,$Y_2$), ‘both $N_1$ and $N_2$’: boy king, politician-tycoon. (“Dvandva” compounds)
• SAME/SIMILAR ($X_1$, $Y_2$), ‘$N_1$ and $N_2$ are the same/similar’: zebrafish, piggy bank, string bean, sunflower. This is not reversible, because the function is symmetric; asymmetry arises only through profiling.
• KIND ($X_1$, $Y_2$), ‘$N_1$ is a kind of $N_2$’: puppy dog, ferryboat, limestone. Reversible: seal pup, bear cub (there are other possible analyses as well, perhaps promiscuously)
• SERVES-AS ($Y_2$, $X_1$), ‘$N_2$ that serves as $N_1$’: handlebar, extension cord, farmland, retainer fee, buffer state.
• LOC ($X_1$, $Y_2$), ‘$N_2$ is located at/in/on $N_1$’: sunspot, window seat, tree house, background music, nose hair, donut hole. Reversible: ‘$N_1$ located at/in/on $N_2$’, or, reprofiled, ‘$N_2$ with $N_1$ at/in/on it’: raincloud, garlic bread, inkpad, stairwell, icewater, water bed.[i]
• LOCtemp ($X_1$, $Y_2$), ‘$N_2$ takes place at time $N_1$’: spring rain, morning swim, 3 a.m. blues. A special case of LOC ($X_1$, $Y_2$).
• CAUSE ($X_1$, $Y_2$), ‘$N_2$ caused by $N_1$’: sunburn, diaper rash, knife wound, surface drag.
• COMP ($Y_2$, $X_1$), ‘$N_2$ is composed of $N_1$’: felafel ball, rubber band, rag doll, tinfoil, brass instrument. Reversible: ‘$N_1$ is composed of $N_2$’, or, reprofiled, ‘$N_2$ that $N_1$ is composed of’: wallboard, bathwater, brick cheese, sheet metal.
• PART ($X_1$, $Y_2$), ‘$N_2$ is part of $N_1$’: apple core, doorknob, fingertip, stovetop, mold cavity. Reversible: ‘$N_2$ with $N_1$ as a part’: snare drum, lungfish, string instrument, ham sandwich, wheelchair. If $N_1$ is a mass noun, this relation paraphrases better as ‘$N_2$ is composed in part of $N_1$’: gingerbread, cinnamon bun, cheesecake, noodle soup. Reversible: ‘$N_2$ that forms part of $N_1$’: stew beef, cake flour, lunch meat.[ii]
• MAKE (X, Y, FROM Z), ‘X makes Y from Z.’ This creates two families of compounds, depending on which two arguments are mapped to $N_1$ and $N_2$.
- ‘$N_2$ made by $N_1$’: moonbeam, anthill, footprint, horse shit. Reversible: ‘$N_2$ that makes $N_1$’: honeybee, lightbulb, musk deer, textile mill
- ‘$N_2$ made from $N_1$’: apple juice, olive oil, grain alcohol, cane sugar, cornstarch. Reversible: ‘$N_2$ that $N_1$ is made from’: sugar beet, rubber tree.[iii]
• PROTECT (X, Y, FROM Z), ‘X protects Y from Z.’ This is the one function in the group that does not seem especially “basic.” It too creates two families of compounds.
- ‘$N_2$ protects $N_1$’: chastity belt, lifeboat, safety pin
- ‘$N_2$ protects from $N_1$’: mothball, flea collar, cough drop, mosquito net, sun hat
The basic functions and the action modalities can fill in F in the modifier schema above to build compound meanings, as in:
$window_1 \space seat_2 = {SEAT_2}^\forall$ ; [LOC ($\forall$ AT $WINDOW_1$)]
$felafel_1 \space ball_2 = {BALL_2}^\forall$; [COMP ($forall$, $FELAFEL_1$)]
(Additional schemas are described in Jackendoff’s paper for more remote cases.)
We can extend this model from abstract conceptual semantics to a formal ontology schema by identifying the domains of the NP constituents. It appears that a preference rule system can express the rule type inference set with good fidelity to what a human curator would produce.
Output is in the form of two kinds of assertions:
- (New) terms that are probable concept names (or synonyms) and that are either common names or named entities, and
- Inferred relationships, possibly involving an existing bootstrap ontology concept, along with zero or more types each with an indication of confidence.
Automatic incorporation of these assertions into an existing ontology is straightforward but should be reviewed by a domain expert.
NPN compounding patterns may also be inferred, though less informative for new ontological inferences from book index material. The forms are very rich as illustrated below.
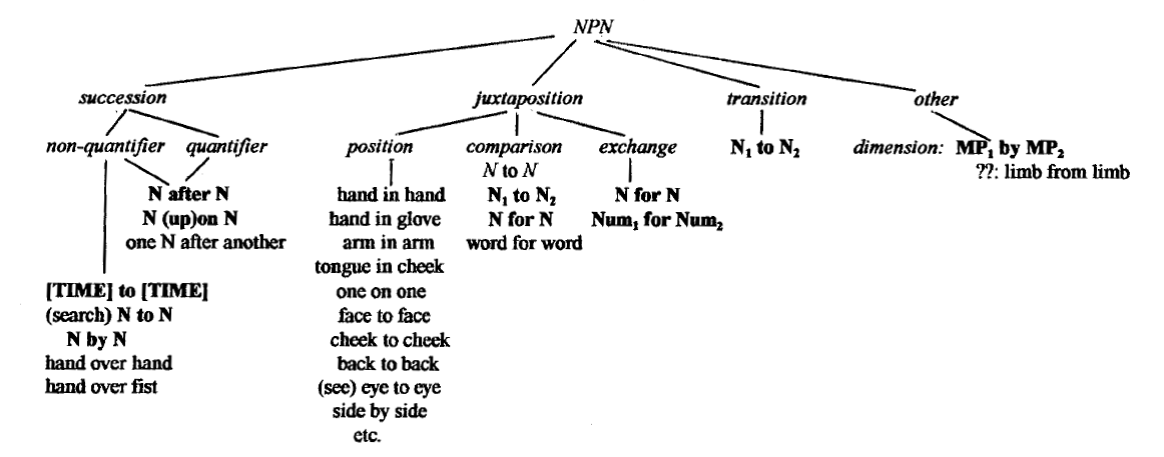
Taxonomy of NPN from Jackendoff, 'Construction after Construction'
- iThese cases verge closely on "X with Y as a part", below. It is not clear to me whether they are distinct.
- iiThe difference between COMP and PART can be illustrated by the ambiguity of clarinet quartet. On the COMP reading it means "quartet of four clarinets"; on the PART reading it means "quartet of which a clarinet is a distinctive member", e.g. a clarinet and three strings.
- iiiThis relation differs from PART in that Z is no longer identifiable as such in Y. However, the distinction is slippery.